Inflammatory Bowel Disease (IBD) Biomarkers Programme


The main types of inflammatory bowel disease (IBD) are Crohn's Disease (CD) and Ulcerative Colitis (UC). These cause considerable suffering to millions of people across the world with numbers of those affected rising rapidly. However, the reasons these diseases arise are poorly understood.
Currently, our knowledge of the causes of IBD are so poor that there are no cures for IBD. Treatments generally focus on clinical management of patient's symptoms and, in severe cases, surgical removal of damaged bowel sections.
IBD-BIOM addresses the urgent need to improve our understanding of IBD. This project is a large EU-funded collaborative research programme to find reliable biomarkers of inflammatory bowel disease. Discovery of such biomarkers should further our understanding of the molecular dysfunctions that give rise to IBD. Our aim is to use such biomarkers in the development of better early-warning clinical diagnostic tests for IBD patients - so they can be given appropriate healthcare and social support promptly and efficiently. This should reduce the suffering of patients and their families and lower the long-term healthcare costs for managing IBD patients.
The IBD-BIOM Research Programme
IBD-BIOM is a European FP7 collaborative scientific research programme for the discovery of diagnostic and prognostic biomarkers for inflammatory bowel disease (IBD).
The programme is EU FP7 project # 305479 and it falls within the FP7 HEALTH.2012.2.4.5-2 theme (Biomarkers and diagnostics for chronic inflammatory diseases of the joints and/or digestive system).
The ten partner organisations in the programme include specialist scientific and medical laboratories across Europe and the US in the areas of clinical treatment of IBD patients, genomics, glycomics and planning and statistical analysis of large-scale biochemical and clinical datasets. The IBD-BIOM research work is planned to span four years (from October 2012 to September 2016) with EU funding of six million Euros.
Aim
Our aim is to discover new, more reliable clinical biomarkers for IBD to:
- Allow the early diagnosis of patients with IBD and
- Point to possible molecular targets for new, improved therapies to alleviate the suffering of IBD patients.
Philosophy
Our philosphy is one of strong cooperation within the IBD-BIOM consortium and with other scientific and medical organisations involved with complementary research on IBD. We also place a high importance on keeping in close touch with people affected by inflammatory bowel disease including patients with IBD, their families and those who care for them. This helps us to keep our focus on producing research outcomes to significantly improve the lives of IBD patients.
IBD-BIOM Consortium
The IBD-BIOM research collaboration partner organisations and principal investigators are as follows:
| Participant no. | Name of Principal Investigator | Participant Legal Name | Short Name | Country | Organisation Type |
|---|---|---|---|---|---|
| 1 | Prof Jack Satsangi | University of Edinburgh | UEDIN | United Kingdom | Academic / Coordinator |
| 2 | Prof Gordan Lauc | Genos Ltd | Genos | Croatia | SME |
| 3 | Dr Daryl L. Fernandes | Ludger Ltd | Ludger | United Kingdom | SME |
| 4 | Dr. Iain Pemberton | IP Research Consulting SAS | IPRC | France | SME |
| 5 | Dr. Vito Annese | University Hospital Careggi, Florence | UHCF | Italy | Hospital |
| 6 | Dr. Manfred Wuhrer | Leiden University Medical Center | LUMC | Netherlands | Academic |
| 7 | Prof Vlatka Zoldos | University of Zagreb | UNI-ZG | Croatia | Academic |
| 8 | Dr. Daniel Kolarich | Max Planck Institute Berlin | MPI | Germany | Academic |
| 9 | Dr. Dermot McGovern | Cedars Sinai Medical Center | CS-MC | United States | Hospital |
| 10 | Dr. Marieke Pierik | Maastricht University Medical Centre | UM | Netherlands | Academic |
Summary of the IBD-BIOM Research Programme
This programme is for development of early warning diagnostics and molecular biomarker discovery for inflammatory bowel disease (IBD) using integrated 'omics' technologies. The IBD-BIOM consortium partners are a multidisciplinary team of leading academic and industrial researchers in IBD, genomics, glycomics, activomics, bionalytical services and diagnostic kits.
IBD affects 2.5 million Europeans and is rising, particularly in children who suffer aggressive forms. Symptoms include irreversible bowel damage, severe diarrhoea, abdominal pain, blood loss from the bowel and uncontrolled leakage of faeces. Current diagnostic tools are poor and treatments limited to symptom relief and surgical removal of bowel sections.
Recent genome-wide association studies (GWAS) performed by IBD-BIOM partners have identified around 100 IBD susceptibility loci, but clinical application has been limited. A promising new approach was shown in 2010 when four partners applied GWAS plus glycomics to diabetes. This led to a patent (UK 1016464.8) for antennary fucose as a highly discriminative glycan biomarker of HNF1A-MODY diabetes.
Serum glycosylation is also altered in inflammation and immune disturbances, so in IBD-BIOM we propose to apply GWAS-glycomics, expanded to include epigenetics and activomics, to the study of IBD patients. Workpackages include development of an IBD biomarker discovery and assay system (IBD-BDAS) that integrates modules for patient cohorts and samples, biochemical profiling by genomics, glycomics and activomics, and statistical analysis of the large datasets. We plan to implement an ISO 9001 quality system to cover IBD-BDAS modules and key proceses would be validated following ICH Q2(R1) and cGxP.
The IBD-BDAS will be used on biological samples from 6000 very well characterised IBD patients and controls available to our EU and US clinical partners. The aim is to discover reliable biomarkers that could allow insights into the molecular pathogenesis of IBD and development of prognostic tools and targeted therapies.
Exploitation includes use of the IBD-BDAS for discovery of useful biomarkers for IBD risk and progression and a clinical diagnostic service for early identification of IBD in susceptible patients.
OUR SERVICES

Inflammatory Bowel disease
Delaying the onset of the disease as well as finding novel potential targets for intervention strategies would dramatically increase the likelihood of healthy, active and independent lives until old age
Inflammatory bowel diseases are characterized by severe chronic intestinal inflammation with huge impact on quality of life. Current treatment is expensive and lacks in therapeutic precision. This reduces efficacy and safety and the disease progress throughout life as well as causes un-necessary side effects from patients receiving treatment they do not respond to. Delaying the onset of the disease as well as finding novel potential targets for intervention strategies would dramatically increase the likelihood of healthy, active and independent lives until old age.
How inflammatory bowel disease is manifested depend on the area of the intestinal tract involved. Many of the symptoms, however, are not specific for this disease. The World Gastroenterology Organization indicates the following symptoms may be associated with inflammatory damage in the digestive tract
• Diarrhea
• Bowel movement abnormalities:
• Abdominal cramping and pain:
• Nausea and vomiting:
If you suspect yourself of having IBD please consult your local physician.
Patients included in this study are recruited via the normal admission to hospitals within the regions where project partners are active. No recruitment will be made outside those regions.
Protein profiling
Proseek Multiplex – measure 92 biomarkers in 1µl of sample without compromising data quality
To enable identification and understanding of molecular and cellular pathways involved in initiation of the diseases we will develop novel and improved methods for large scale sensitive affinity proteomics based on partner Olink’s proprietary Proximity Extension Assay (PEA) technology. Initially developed in the FP7 program Proactive it is now made available and sold under the trade name Proseek Multiplex. In this project we will build dedicated multiplexed panels targeting proteins of interest for IBD.
Our hypothesis is that (i) large numbers of protein biomarkers must be screened in order to find the best panels of relevant diagnostic markers, (ii) individual protein biomarkers will not be sufficient as diagnostic marker and (iii) informative tissue leakage protein biomarkers will be present at low concentration in circulation. Current multiplexed protein formats are limited either by their low sensitivity, low throughput or applicability for clinical testing, or all three. Therefore we aim to utilize Proseek multiplex, a multiplexed protein assay capable of detecting low abundant protein biomarkers in high throughput format.
Proseek Multiplex is based on the PEA developed at Olink. PEA has a major advantage over conventional multiplex immunoassays in that only correctly matched antibody pairs give rise to a signal. PEA is a homogeneous assay that uses pairs of antibodies equipped with DNA reporter molecules. When binding to their correct targets, they give rise to new DNA amplicons each ID-barcoding their respective antigens. The amplicons are subsequently quantified by high throughput real-time PCR.
Proseek® Multiplex can provide accurate quantification of up to 92 proteins in one run at low to sub picogram per milliliter levels from1 µl samples such as serum or tissue lysates.
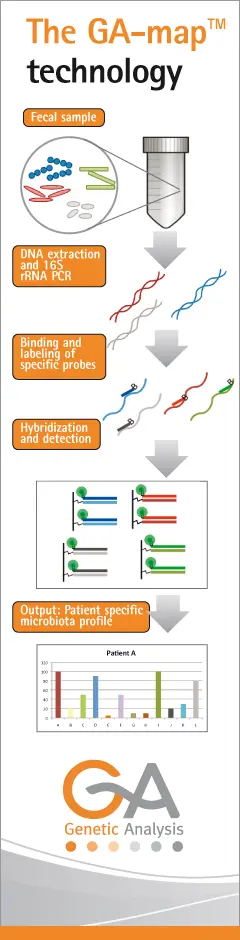
Microbial profiling
The GA-map allows screening of many samples using a panel tailor-made for IBD analysis
It is now well recognized that understanding microbial content of the gut is relevant for inflammatory bowel diseases . However little is known about the aetiology – if changes in microbial content of the gut are causative or an effect of disease progression. Furthermore there are lacking large collaborative studies coupling knowledge about gut microbial content and host genetic factors as well as identification of differences in gut microbiota early on in the IBD. Therefore, a comprehensive study of the gut microbiota will be important for this project. In order to allow screening of large numbers of patients for presence of many different microbes, we will develop an IBD-relevant microbe bead-array (called GA-map™). By performing 16S rRNA amplicon pyro-sequencing of collected samples from IBD patients and healthy controls, a catalogue of microbes of interest in IBD will be established. This in combination with other public projects for microbe cataloguing, e.g. and the Human Microbiome project and MetaHIT , partner GA will design unique and specific DNA probes for the GA-map platform, capable of reporting the presence of the various selected microbes. Based on the observations that there is high intra-individual variation of intestinal microbiomes in the human population, we hypothesize that characterizing IBD microbiomes could disclose bacterial and fungal species implicated in IBD aetiology. To better understand how microbiomes relate to human disease, we will undertake a comparative sequencing survey of the microbiomes present in faces in 40 IBD patients and 40 unaffected individuals using whole metagenomic sequencing in order to verify data from GA-map and look for additional microbial markers outside the 16sRNA amplicon.
Currently pyro-sequencing of the 16S rRNA gene is the most commonly used method for studying diversity in the gut microbiota . Although the method is useful as a discovery tool, the output of the sequence analysis requires several complex computational steps, as well as some subjective decisions as to sequence identity. Reducing the number of samples in each run will reduce the complexity of analysis, but this will drastically increase the cost per sample. Therefore, in order to be able to do a quality comparison of all samples, a novel technology (GA-map™) will be adapted that utilizes a combination of specific DNA probes to profile the gut microbiota. The GA-map™ technology platform is also included in another ongoing FP7 project (KBBE.2011.2.2-01, grant no 289517), and will here be further developed and tailor-made for IBD analysis. To our knowledge, this will be the first time a microbial profile will be obtained and compared from this many clinical samples. In addition, whole genome metagenomic sequencing of a sub-set of samples is expected to give additional information on biological markers and phenotypic traits of important bacteria contributing to disease. These various microbial profiling approaches will be performed on a unique sample material, namely from patients just diagnosed with IBD and before any treatment has commenced. Using this unique material will help us to understand the mechanism of initiation of disease, as well as identify markers or microbial profiles that can be used for early diagnosis of disease.
Nucleic Acid analysis
Large scale analysis of methylation patterns and transcription profiles will be conducted to complement traditional genotyping, given the low contribution of found genetic alternations to explain for heritability of IBD
Recent advances made by large international consortia, with active contribution of members in this project, have identified a number of susceptibility loci in both ulcerative colitis (UC) and Crohn’s disease (CD) , Follow up studies of > 50000 cases and controls, have lead to that now 163 genes/loci and associated SNPs have been confirmed and recently identified as true IBD (CD or UC) genetic susceptibility factors . The project will score information of already existing susceptibility genotypes as a “baseline” for the further analysis and complement the analysis with methylation and transcription profiling within this unique cohort of patients.
The contribution of epigenetic alterations to disease pathogenesis is now emerging as a research priority given the low contribution of found genetic alternations to explain for heritability. To advance the understanding of epigenetic changes in IBD we will perform large scale methylation profiling. The means to analyze and interpret data from large scale methylation studies is still in its early phases but the size of the cohort have been chosen accordingly based on recommendations in recent publications to allow relevant data to be extracted. . The current study will generate the largest methylation profiling in early IBD patients so far. Partners within the project have recently found epigenetic factors relevant for development of CD also in blood. This is a very interesting finding as it opens up for blood based screening of biomarkers as well as explore if epigenetic regulation may represent a target for therapeutic intervention, and it will be further pursued within the project. One of the project aims is to understand the mechanism for initiation of the disease and therefore methylation profiling will also be performed on mucosal biopsies of the actual disease relevant issue.
To complement the protein assays, as described in the section about proteins, and to facilitate interpretation of the methylation data we will also perform RNA expression analysis on the entire cohort. The RNA expression will be analyzed using high content RNA expression arrays to yield a global view of gene regulation changes in the early stages of IBD.
In addition to array based molecular phenotyping we will perform in depth analysis of a subset of the cohort. Based on the initial diagnose of the IBD patients they will be divided in patients with severe initial disease and more mild symptoms and on a subgroup we will perform whole genome sequencing of bi-sulfite concerted DNA to yield a complete map of methylation changes. The same patients and samples will be subjected to Whole transcriptome Shotgun Sequencing (WTSS) to yield a complete map of expression as well as RNA variants.